Introduction
One of the strengths of Python, and specifically Cartopy, is that it's pretty easy to combine many different datasets with varying geographic coordinates onto a single map. In this tutorial, I'll combine background "Blue and Black" marble images from NASA with the forecast aurora data from the Space Weather Prediction Center (SWPC:
https://www.swpc.noaa.gov/), and simulated cloud cover from the GFS to produce an aurora forecast map. Now, since one of my main goals for this blog is not to simply regurgitate what already exists, if you are just looking to plot the aurora forecast, there is an excellent example (which I'll be building off of in this tutorial) of how to so in the Cartopy gallery:
here.
This is a somewhat advanced tutorial, so I won't be focusing on trying to explain a lot of the plotting minutia or reasoning. If you're just starting out it is recommended that you check out some of my beginner tutorials first:
Introduction to Cartopy, and
my post on how to read in NetCDF data.
In this tutorial you will learn how to:
- Load background images into Cartopy
- Use the Cartopy "Nightshade" feature to blend the NASA blue and black marble images
- Maskout shapefile geometries
- Load the aurora forecast from the SWPC using Numpy "loadtxt"
- Create a custom colormap using matplotlib "LinearSegmentedColorMap"
- Use the NearsidePerspective projection from Cartopy
The end result of the tutorial will be an image that looks similar to this, but will be different depending on the time of day, auroral activity, and cloud cover:
 |
| Final Product: Forecast Aurora Probability and simulated GFS clouds valid UTC time. |
|
|
|
|
Loading the background images
The first step is to download the "Blue Marble" and "Black Marble" background images to load into the dataset. These are pretty easy to grab, I will simply provide links to the geotiff 0.1 degree versions for each image. Load these into a folder where you can access them using a Python script.
If these don't work you can try the parent sites:
Blue Marble here, and
Black Marble here.
Now that you have the data, it's time to load it into Python. Before that however, the required packages for the entire tutorial need to be imported:
try:
from urllib2 import urlopen
except ImportError:
from urllib.request import urlopen
from io import StringIO
import numpy as np
from datetime import datetime, timedelta
import cartopy.crs as ccrs
from cartopy import feature as cfeature
import matplotlib.pyplot as plt
from matplotlib.colors import LinearSegmentedColormap
from cartopy.feature.nightshade import Nightshade
from matplotlib.path import Path
from cartopy.mpl.patch import geos_to_path
import netCDF4 as NC
Note the"try" statement is used to make the script compatible with both Python 2.7 and Python 3 urllib syntax. Now, once the data is imported, I'm going to define a common function to load the "Marble" images, since they have identical lat/lon boundaries.
def load_basemap(map_path):
img=plt.imread(map_path)
img_proj = ccrs.PlateCarree()
img_extent = (-180, 180, -90, 90)
return img, img_proj,img_extent
You should be able to see that the above function is really straightforward. The image is first loaded into an array using "imread" from matplotlib, and then the image extent is set to span the entire glob (which is obvious from looking at the image), and then the image projection information for Cartopy is set to PlateCarree, since we are working in lat/lon coordinate space. To investigate a little further, we can call this function and look at the shape of the image array.
Assuming the image is in the same folder as your script...
map_path='RenderData.tif'
bm_img,bm_proj,bm_extent=load_basemap(map_path)
print(np.shape(bm_img))
returns: (1800, 3600, 3)
Notice that the array returned has 3 dimensions despite the fact that it is a 2D image: the 3rd column is where the individual red/green/blue information for the image is stored, where as the first 2 columns correspond to the y and x pixels. To demonstrate this, I'll plot each channel separately without geographic information.
plt.figure(figsize=(8,14))
ax=plt.subplot(311)
plt.pcolormesh(bm_img[:,:,0],cmap='Reds')
ax=plt.subplot(312)
plt.pcolormesh(bm_img[:,:,1],cmap='Greens')
ax=plt.subplot(313)
plt.pcolormesh(bm_img[:,:,2],cmap='Blues')
plt.show()
This produces the following image:
 |
| RGB channels for the Blue Marble image (Yikes! It's upside down!) |
Note that where all three channels are saturated, the combined RGB image will be white (e.g., Antarctica), where as where all three channels are faded, the RGB image will appear back (e.g., the Oceans). Also notice how the image is upside down, this will be addressed in the next part.
The question, is how can we plot the combined RGB image from the 3 dimensional array. Instead of using the "pcolormesh" or the "contourf" functions, the "imshow" function is used. this function, is versitile and naturally takes a 3D array assuming the 3rd axis (if there is one) is the RGB channel. It also, takes the extent argument, which defines the image corners, without needed corresponding coordinate arrays.
plt.figure(figsize=(11,8))
ax=plt.subplot(111,projection=ccrs.PlateCarree())
ax.imshow(bm_img,extent=bm_extent,transform=bm_proj,origin='upper')
plt.show()
This results in the following image:
 |
| Blue Marble |
I won't go into detail on how to load the black marble, however, I will provide the code used to load both images as they are read into the final product.
bm_img,bm_proj,bm_extent=load_basemap('RenderData.tif')
nt_img,nt_proj,nt_extent=load_basemap('BlackMarble_2016_01deg_geo.tif')
Blend Images using the Day/Night Delimiter
This next section is the crux of the whole tutorial: Basically, I'll explain how to use Cartopy Nightshade (Note: requires version 0.17 or higher) to mask out the day time blue marble and show the night time black marble image. For this, I'll use the matplotlib "Path" function to draw a polygon "path" connecting the coordinate information associated with the Nightshade function.
The Nightshade function is a neat function that allows you to draw the day/night delimiter on a glob for a given datetime object. In the full example, the datetime object will be defined by the aurora forecast, but for demonstration of Nightshade, the datetime.now() object will be used.
For a simple demonstration, simply copying the code to generate the blue marble image, with only a couple of added lines, provided you are following the tutorial with the package imports written as above:
plt.figure(figsize=(11,8))
ax=plt.subplot(111,projection=ccrs.PlateCarree())
ax.imshow(bm_img,extent=bm_extent,transform=bm_proj,origin='upper')
ax.add_feature(Nightshade(datetime.utcnow()),zorder=2,alpha=0.8,color='r')
plt.title(datetime.utcnow().strftime('%c'))
plt.show()
This will essentially mask out the "night sky" over the blue marble image, it's that simple!
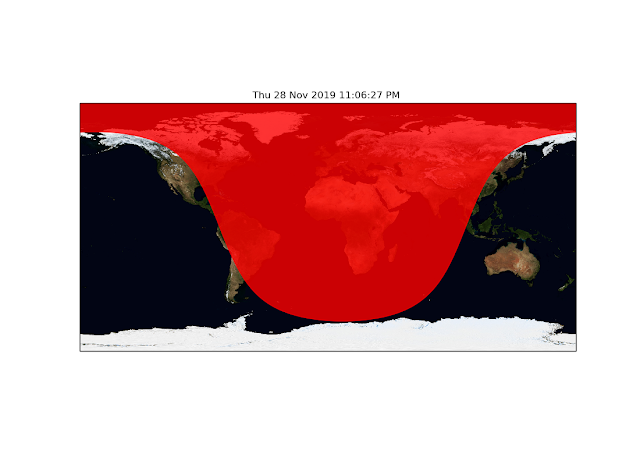 |
| Night masked out. |
Now the goal here is simple: replace the "red" masked out section with the black marble image. To do that, I'll first define a function to clip regions within the shapefile polygon.
The first step is to define the Nightshade "shape" from the Nightshade function:
nshade=Nightshade(datetime.utcnow())
transform=nshade.crs
Now we have both the nightshade shape, and the nightshade coordinate reference system, which is needed to ensure the projection transform is performed correctly. From the shape, we grab the geometries (i.e., the polygons, or in this case ... polygon; singular).
geoms=list(nshade.geometries())
Here is the tricky part: because the Nightshade coordinate reference system is NOT Plate Carree (it's a rotated pole projection for those interested), it's important that the transform is done correctly. If you assume the transform is Plate Carree you will end up with a mask that is wrong. However, the matplotlib path function requires that the transform is a matplotlib transform, and not a cartopy crs object. So, to do this conversion, I will define a "dummy" transform when defining my Caropy subplot that equals the crs from the nightshade object.
fig = plt.figure(figsize=(10, 8))
ax = fig.add_subplot(1, 1, 1, projection=ccrs.PlateCarree(),transform=nshade.crs)
Now, we have a defined figure, and a matplotlib transform that matchs the nightshade object, we're ready to mask. First we plot the blue and black marble images on the same subplot, and then use the geos_to_path and Path functions imported at the top of the script:
plt.figure(figsize=(11,8))
ax=plt.subplot(111,projection=ccrs.PlateCarree(),transform=nshade.crs)
im0=ax.imshow(bm_img,extent=bm_extent,transform=bm_proj,origin='upper',zorder=1)
im1=ax.imshow(nt_img,extent=nt_extent,transform=nt_proj,origin='upper',zorder=2)
path = Path.make_compound_path(*geos_to_path(geoms))
im1.set_clip_path(path, transform=ax.get_transform())
plt.title(datetime.utcnow().strftime('%c'))
plt.show()
And the blended image is complete,note that the subplot transform is used in the set_clip_to_path function:
 |
| Blended image based on current datetime. |
Now, it's time to plot the aurora forecast.
Loading and Plotting the Aurora Forecast
This section will mostly be a reproduction of the code demonstrated
here, with some minor differences. Esentially, Numpy loadtxt is used to load the ascii data from the SWPC into a 2 dimensional array and the array is plotted on the map. The first step is to define a cool looking aurora color bar. This a a
direct reproduction from the Cartopy example because I think they did a nice job with the color scales.
def aurora_cmap():
"""Return a colormap with aurora like colors"""
stops = {'red': [(0.00, 0.1725, 0.1725),
(0.50, 0.1725, 0.1725),
(1.00, 0.8353, 0.8353)],
'green': [(0.00, 0.9294, 0.9294),
(0.50, 0.9294, 0.9294),
(1.00, 0.8235, 0.8235)],
'blue': [(0.00, 0.3843, 0.3843),
(0.50, 0.3843, 0.3843),
(1.00, 0.6549, 0.6549)],
'alpha': [(0.00, 0.0, 0.0),
(0.70, 1.0, 1.0),
(1.00, 1.0, 1.0)]}
return LinearSegmentedColormap('aurora', stops)
Basically, a dictionary with 0-1 values for red (r), green (g), blue (b), and transparancy (alpha) is passed to the LinearSegmentedColormap function which defines a color map that basically linearly interpolates between each of the 9 "stops" defined by the user.
Now, that the colormap is defined, you can pull the aurora data from the SWPC and load it into an array:
# To plot the current forecast instead, uncomment the following line
url = 'http://services.swpc.noaa.gov/text/aurora-nowcast-map.txt'
response_text = StringIO(urlopen(url).read().decode('utf-8'))
aurora_prob = np.loadtxt(response_text)
# Read forecast date and time
response_text.seek(0)
for line in response_text:
if line.startswith('Product Valid At:', 2):
dt = datetime.strptime(line[-17:-1], '%Y-%m-%d %H:%M')
Now, again this code is largely copied from the Cartopy example, but essentially you're reading the text information from the URL, first into the Numpy loadtxt function, which is a natural fit for this data, since the function, by default, assumes "#" is a comment, and "spaces" are delimiters, so in this instance, no additional arguments are required for loadtxt. Then, the datetime associated with forecast is read into a datetime object.
Since it is known that the lat/lon data spans -90 to 90 and -180 to 180, we define lat/lon arrays based on the shape of the aurora data:
lons=np.linspace(-180,180,np.shape(aurora_prob)[1])
lats=np.linspace(-90,90,np.shape(aurora_prob)[0])
Finally, combining everything together to product the "cloud-free" aurora forecast:
plt.figure(figsize=(11,8))
ax=plt.subplot(111,projection=ccrs.PlateCarree(),transform=nshade.crs)
im0=ax.imshow(bm_img,extent=bm_extent,transform=bm_proj,origin='upper',zorder=1)
im1=ax.imshow(nt_img,extent=nt_extent,transform=nt_proj,origin='upper',zorder=2)
path = Path.make_compound_path(*geos_to_path(geoms))
im1.set_clip_path(path,transform=ax.get_transform())
Z=ax.contourf(lons,lats,np.ma.masked_less(aurora_prob,1), levels=np.linspace(0,100,61), transform=ccrs.PlateCarree(), zorder=5,cmap=aurora_cmap(),antialiased=True)
plt.title(datetime.utcnow().strftime('%c'))
plt.show()
Which gives you the following image:
 |
| Aurora Forecast with Plate Carree Projection |
Finally, replacing the PlateCarree map projection with the NearsidePerspective projection, we get something a little bit closer to the final image:
fig = plt.figure(figsize=[10, 8])
ax = plt.axes(projection=ccrs.NearsidePerspective(-170, 45),transform=nshade.crs)
im0=ax.imshow(bm_img,extent=bm_extent,transform=bm_proj,origin='upper',zorder=1)
im1=ax.imshow(nt_img,extent=nt_extent,transform=nt_proj,origin='upper',zorder=2)
path = Path.make_compound_path(*geos_to_path(geoms))
im1.set_clip_path(path, transform=ax.get_transform())
Z=ax.contourf(lons,lats,np.ma.masked_less(aurora_prob,1), levels=np.linspace(0,100,61), transform=ccrs.PlateCarree(),
zorder=5,cmap=aurora_cmap(),antialiased=True)
plt.title("Forecast Aurora Probabilty \n Valid: %s"%dt.strftime('%b/%d/%Y %H:%M UTC'),
loc='left',fontweight='bold')
 |
| Orthographic / Nearside Perspective Projection |
Adding A Cloud Forecast from the GFS
The final step in this tutorial is to add cloud cover from the GFS to the map. This is unnecessary, if you're happy with the above image, but my personal thought is, you aren't going to see the aurora if it's cloudy, so having the forecast cloud cover is a nice overlay. It's real easy to add the clouds, simply pull the GFS data from the
NOMADS server using netCDF4, match the GFS model time to the aurora forecast time and overlay the total cloud cover variable on the map. Again, my assumption is that if you've made it this far, you're comfortable working with GFS data using netCDF4 and I won't explain in detail why the following code works:
dhour='12' ## or 00, or 18 or 06, your choice
dtstr=dt.strftime('%Y%m%d')
gfs_path='http://nomads.ncep.noaa.gov:80/dods/gfs_0p25_1hr/gfs%s/gfs_0p25_1hr_%sz'%(dtstr,dhour)
gfs_data=NC.Dataset(gfs_path,'r')
gfslons=gfs_data.variables['lon']
gfslats=gfs_data.variables['lat']
tres=gfs_data.variables['time'].resolution
tmin=datetime.strptime(gfs_data.variables['time'].minimum,'%Hz%d%b%Y')
times=[timedelta(hours=float(i)*24.*tres)+tmin for i in range(len(gfs_data.variables['time'][:]))]
tidx=np.argmin(np.abs(np.array(times)-dt))
clouds=gfs_data.variables['tcdcclm'][tidx,:]
and then simply add the following code to the beneath the code used to load the background images and plot the aurora forecast:
ax.pcolormesh(gfslons[:],gfslats[:],np.ma.masked_less(clouds,75.),transform=ccrs.PlateCarree(),
alpha=0.7,cmap='Greys_r',zorder=4,vmin=50,vmax=100)
And voila:
 |
| Final Product |
Final Notes
That is all for this tutorial, it was a long one, but I think there is a lot of useful stuff in here. A few final notes, reprojecting the background images can lead to really poor quality images, I find this particularly true for Lambert and PolarStereo projections, so be aware of that.


















